| Name |
ring finger protein 2 |
SBDS ribosome maturation factor |
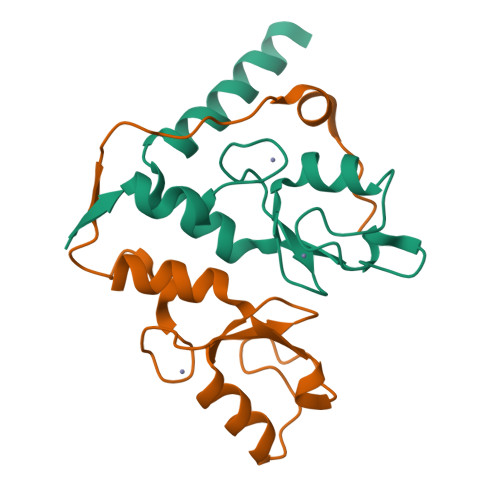
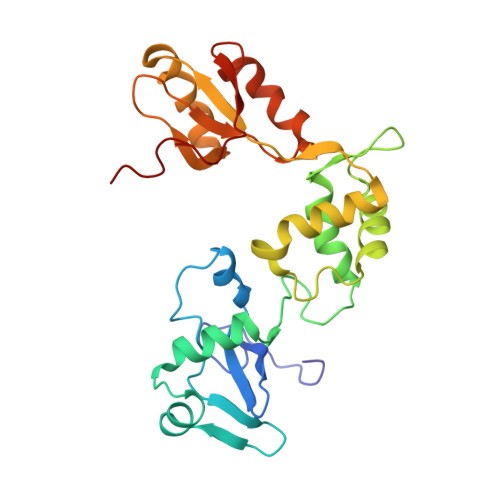
| Structure |
|
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
None
|
| Diseases |
None
|
|
| GWAS |
- Clopidogrel active metabolite levels ( 28207573
)
- Itch intensity from mosquito bite adjusted by bite size ( 28199695
)
- Obesity-related traits ( 23251661
)
|
No GWAS studies recorded
|
| Interacting Genes |
65 interacting genes:
ABCB1
AMBRA1
BAALC
BMI1
CASP3
CASP9
CBX4
CBX6
CBX7
CBX8
CIP2A
DYSF
ERG
GMNN
H2AC18
H2AC20
H2AC4
H2BC3
H3-4
KAT8
KMT2A
MBD1
MTNR1A
OTUD6A
PCGF1
PCGF2
PCGF6
PGP
PHB2
PHC1
PHC2
PLK1
PSMC4
RING1
RRM1
RYBP
SBDS
SCMH1
SIK1
SMURF2
SSX2
TARDBP
TFCP2
TMEM132D
TSC22D2
UBC
UBE2D1
UBE2D2
UBE2D3
UBE2D4
UBE2E1
UBE2E2
UBE2E3
UBE2H
UBE2J1
UBE2K
UBE2L3
UBE2U
UBE2V1
UBE2V2
UBE2W
UBE3A
USP11
USP19
WASHC1
|
10 interacting genes:
ACD
CEBPA
CLN3
EFL1
KLHL6
LMNA
POT1
RNF2
TERF1
TERF2IP
|
| Entrez ID |
6045 |
51119 |
| HPRD ID |
07028
|
07393
|
| Ensembl ID |
ENSG00000121481
|
ENSG00000126524
|
| Uniprot IDs |
Q99496
|
A0A0S2Z5I7
Q9Y3A5
|
| PDB IDs |
2H0D
3GS2
3H8H
3IXS
3RPG
4R8P
4S3O
6WI7
6WI8
7ND1
8GRM
8PP7
9DBY
9DDE
9DGG
|
2KDO
2L9N
5AN9
5ANB
5ANC
6QKL
|
| GO Terms Enriched among Interactors |
|
|