| Name |
MRE11 double strand break repair nuclease |
ATR checkpoint kinase |
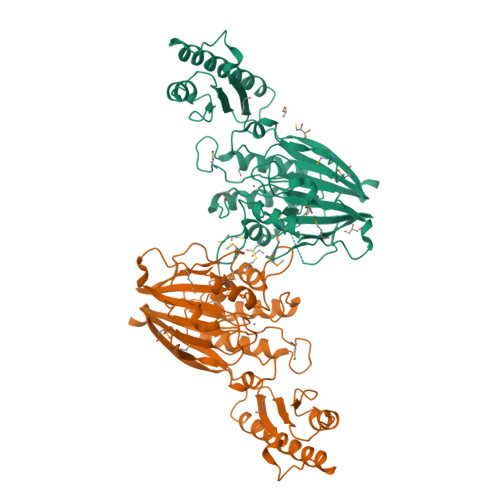
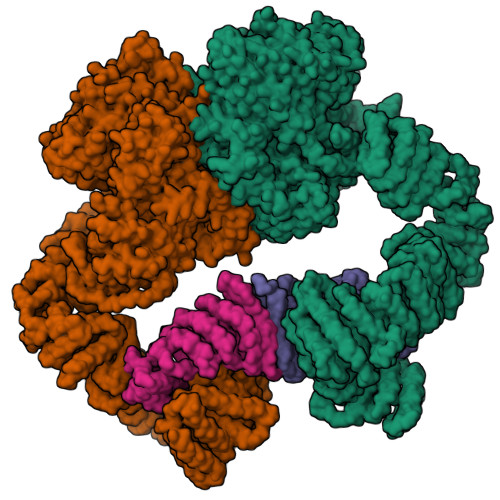
| Structure |
|
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
|
| Drugs |
None
|
|
| Diseases |
|
|
| GWAS |
No GWAS studies recorded
|
|
| Interacting Genes |
27 interacting genes:
ATM
ATR
CCNE1
CDK2
CDX2
CEBPA
DCLRE1C
DYNLL1
EP300
FANCD2
GRB2
H1-2
H2AX
H4C1
LIG1
LOX
MAPK8IP2
NBN
NEK1
NKX3-1
PRKDC
RAD50
RECQL5
SPOP
SUMO1
SUMO2
XRCC6
|
58 interacting genes:
AATF
ABL1
AP1B1
AP3B1
APBB1
ARHGEF1
ATM
BLM
BRCA1
BRCA2
CDKN2C
CEP164
CHD4
CHEK1
CHEK2
CHUK
CLSPN
CREB1
DCAF1
DCLRE1C
DTL
E2F1
E4F1
EEF1E1
EP300
ETAA1
ETV1
FANCA
FANCD2
FANCI
FLT1
H2AX
KDR
LIG4
MCM2
MCPH1
MRE11
MSH2
NBN
NFE2L2
PA2G4
PARP1
PIK3CA
POLD1
POLN
PPP2R3A
RAD17
RASSF1
RHEB
RPA1
TP53
TREX1
UHRF1
UHRF2
UPF1
USP2-AS1
XPA
XRCC5
|
| Entrez ID |
4361 |
545 |
| HPRD ID |
02889
|
08369
|
| Ensembl ID |
ENSG00000020922
|
ENSG00000175054
|
| Uniprot IDs |
F8W7U8
P49959
Q05D78
|
Q13535
|
| PDB IDs |
3T1I
7ZQY
8BAH
8K00
|
5YZ0
|
| GO Terms Enriched among Interactors |
|
|