| Name |
hydroxysteroid 17-beta dehydrogenase 14 |
PHD finger protein 1 |
| Structure |
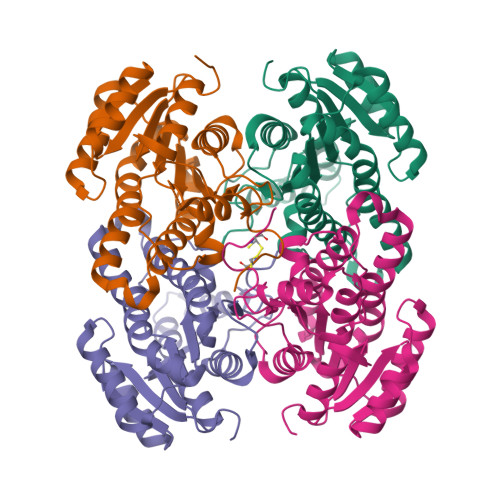
|
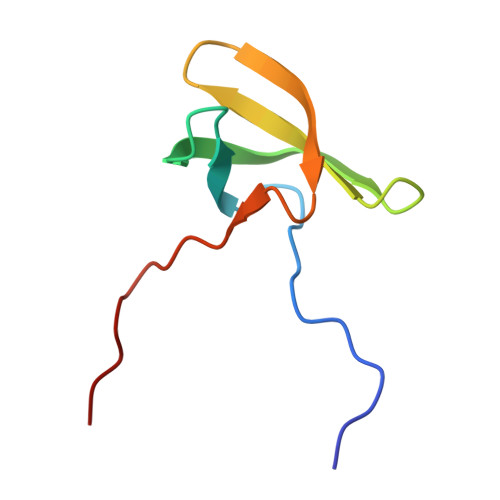
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
|
| GWAS |
|
- Autism spectrum disorder or schizophrenia ( 28540026
)
- Body mass index ( 26426971
)
- Chronic obstructive pulmonary disease or resting heart rate (pleiotropy) ( 30940143
)
- Refractive error ( 32231278
)
|
| Interacting Genes |
24 interacting genes:
AKIRIN2
BRIP1
CA8
CDKN2D
CYTOR
DDIT3
INCA1
MIA3
MIR4435-2HG
MPG
NEK6
NTAQ1
NUDT18
OTUD3
PGLYRP3
PHF1
PICK1
PIDD1
PSMA1
REL
SNAPC3
SNRPC
SREK1IP1
TBC1D22B
|
77 interacting genes:
ABI2
ARID5A
ARRDC3
ATXN7L1
AVPI1
BHLHE40
BIRC7
BOLL
CALCOCO2
CSTPP1
DISC1
DPF1
DYSF
EFHC2
EZH2
FHL2
FOSB
GOLGA6L9
GPKOW
GPRASP3
H3C1
H3C10
H3C11
H3C12
H3C2
H3C3
H3C6
H3C7
H3C8
H4C1
HOMEZ
HSD17B14
INCA1
IRAK1BP1
KIAA0408
KRT40
LBX1
LZTS2
MAGED1
MDFI
MEOX2
MORN3
NAB2
NEK6
PATZ1
PDLIM7
PFDN5
PIBF1
PRR20A
PRR20B
PRR20C
PRR20D
PRR20E
RBBP8NL
RBPMS
SMG9
SORBS3
SPAG8
TFCP2
TFIP11
THAP1
TLE5
TNS2
TOX2
TP53BP2
TRIM23
USP54
VAC14
ZBTB10
ZBTB16
ZIM2
ZNF500
ZNF526
ZNF688
ZNF71
ZRANB1
ZSCAN32
|
| Entrez ID |
51171 |
5252 |
| HPRD ID |
16801
|
04195
|
| Ensembl ID |
ENSG00000087076
|
ENSG00000112511
|
| Uniprot IDs |
A0A140VJH8
Q9BPX1
|
A0A140VJR4
A0A1U9X8A3
O43189
|
| PDB IDs |
1YDE
5EN4
5HS6
5ICM
5ICS
5JS6
5JSF
5L7T
5L7W
5L7Y
5O42
5O43
5O6O
5O6X
5O6Z
5O72
5O7C
6EMM
6FFB
6G4L
6GBT
6GTB
6GTU
6H0M
6HNO
6QCK
6ZDE
6ZDI
6ZR6
6ZRA
6ZT2
|
2E5P
2M0O
4HCZ
5XFN
5XFO
5XFP
6WAT
6WAV
7LKY
8H43
|
| GO Terms Enriched among Interactors |
None
|
|