| Name |
bystin like |
midline 1 |
| Structure |
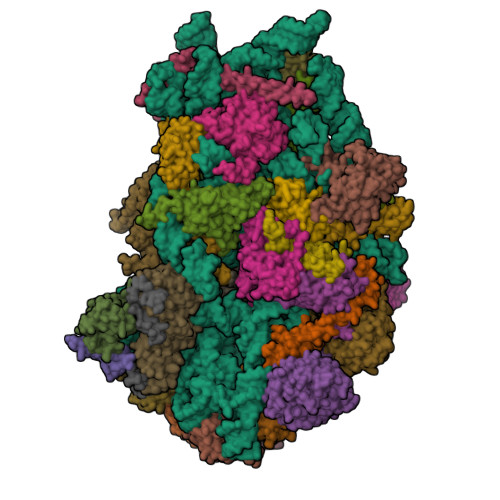
|
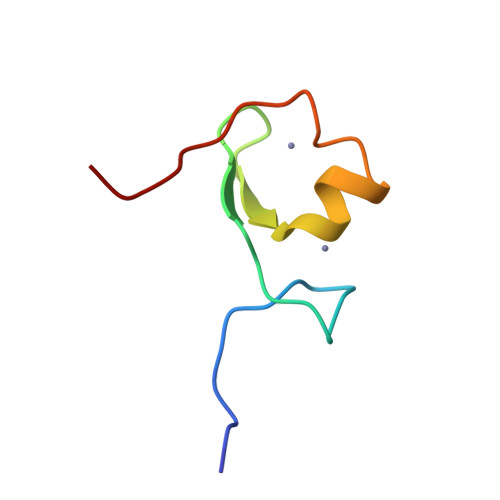
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
|
| Diseases |
None
|
|
| GWAS |
|
No GWAS studies recorded
|
| Interacting Genes |
138 interacting genes:
AIMP2
AMOTL2
APP
ATP5F1B
AXIN2
BEND7
BFSP1
BHLHE40
C1orf94
CAVIN4
CCDC102B
CCDC136
CCDC33
CDC23
CDCA7L
CEP44
CEP57L1
CEP70
COIL
DDX17
DOCK8
DRC4
DVL2
EAPP
EIF4ENIF1
EMD
EPS8
FAM228A
FAM9B
FCHO1
FXR1
FXR2
GMCL1
GOLGA2
GOLGA6L9
GRIPAP1
HMBOX1
HOMEZ
HOOK2
HSF2BP
IKZF1
IKZF3
JRK
KATNAL1
KIFC3
KLHL2
KLHL6
KRT31
KRT40
KRT8
KRTAP10-3
KRTAP10-5
KRTAP10-7
KRTAP4-2
L3MBTL3
LDOC1
LHX3
LMNA
LMO1
LMO2
LONRF1
LZTS1
LZTS2
MB21D2
MCIDAS
MEOX1
MEOX2
MID1
MID2
MIPOL1
MKRN1
MRFAP1L1
MTUS2
NECAB2
NF2
OGT
OLIG3
OSBPL3
PDE4DIP
PHC2
PICK1
PIH1D1
PNMA1
PNMA2
PRICKLE1
PSMC6
RACGAP1
RALY
RALYL
RBAK
RP9
RUBCN
SMN1
SMN2
SNW1
SSX2IP
STX11
TBC1D26
TEKT1
TFIP11
THAP1
TLE5
TNIP1
TRAF2
TRAF4
TRAK1
TRIM14
TRIM27
TRIM37
TRIM38
TRIM41
TRIM54
TRIM55
TRIP6
TRO
TROAP
USH1G
USO1
VIM
VPS37B
VPS52
WASF3
WTAP
ZBTB14
ZBTB8A
ZC2HC1C
ZFP64
ZMAT5
ZNF212
ZNF286A
ZNF426
ZNF438
ZNF48
ZNF655
ZNF668
ZNF71
ZNF835
ZSCAN22
|
46 interacting genes:
BYSL
CDC37
CRY2
DYRK4
EHHADH
ELOA
EPN2
FAM50B
FKBP1A
GMCL1
HMG20B
HTT
IGBP1
KIF9
MEOX1
MID1IP1
MID2
N4BP1
OTUB2
PAX6
PDE4B
PKN1
PPM1A
PPP2CA
PPP2CB
PTCD2
PTPA
RIC8A
SPAG5
STK36
TCEANC
UBE2D1
UBE2D2
UBE2D3
UBE2D4
UBE2E1
UBE2E2
UBE2E3
UBE2K
UBE2L3
UBE2L6
UBE2N
UBE2V1
UBE2W
UBTD1
ZNF618
|
| Entrez ID |
705 |
4281 |
| HPRD ID |
04848
|
02047
|
| Ensembl ID |
ENSG00000112578
|
ENSG00000101871
|
| Uniprot IDs |
Q13895
|
A0A087X255
A0A8I5KPE0
A0A8I5KR14
O15344
|
| PDB IDs |
6G18
6G4S
6G4W
7WTT
7WTU
7WTV
7WTW
7WTX
7WTZ
7WU0
|
2DQ5
2FFW
2JUN
5IM8
7QRY
|
| GO Terms Enriched among Interactors |
|
|