| Name |
reticulon 3 |
speckle type BTB/POZ protein |
| Structure |
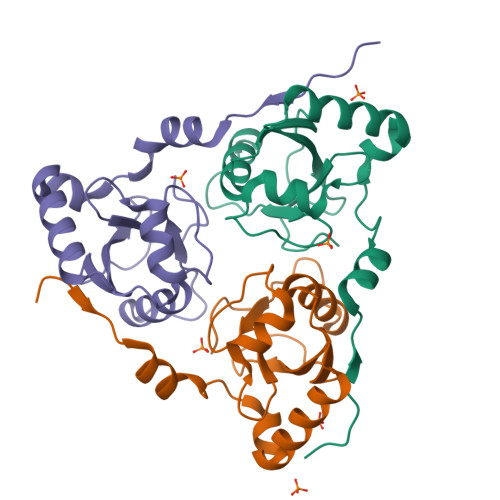
|
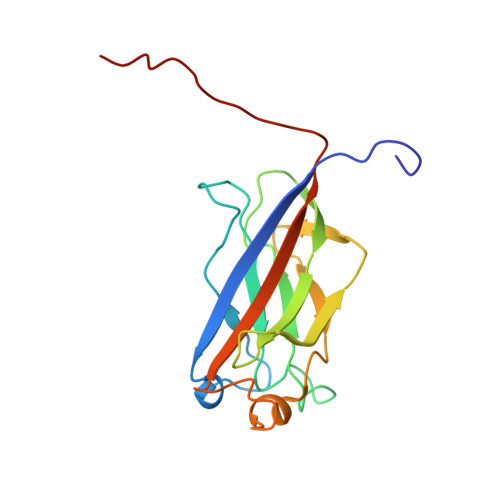
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
|
| GWAS |
No GWAS studies recorded
|
|
| Interacting Genes |
32 interacting genes:
AGTR1
CCR2
CCR8
CDIPT
CERT1
DRD2
FGFR1
FHIP1B
FXR2
GLP1R
GPR152
GPR35
LMNA
MAP1LC3A
MITD1
NDRG4
PLEKHF2
PLN
PPP2R3C
PTPN9
RAB33A
RMDN2
RTN4
SACM1L
SHANK3
SLC30A3
SLC30A8
SNX1
SPOP
TERF1
UGCG
ZNF391
|
74 interacting genes:
AEN
AMOTL2
AR
ATM
BRAF
BRD2
BRD3
BRMS1
CACUL1
CBX4
CCNE1
CDK4
CHAF1A
CHD3
CREB5
CTDSPL2
DAXX
DDIT3
DHX15
DPPA2
DUSP7
EGLN2
ERF
ERG
FAF1
G3BP1
GATAD2A
GCC1
GCC2
GIT2
GLI2
GMEB1
HCST
HIPK2
HMGCS1
ILF3
IRF1
KPNA5
LRCH4
MACROH2A1
MRE11
MYD88
NANOG
NUDCD3
PDPK1
PDX1
PIAS1
PIAS3
PTEN
RANBP9
RBFOX2
RBPJ
RPRD2
RTN3
RXRB
SETD2
SRRM1
SUMO1
TCOF1
TOPORS
TP53BP1
TPD52
TRAF1
TRAF6
TWIST1
UBE2D1
UBE2D2
UBE2E3
UBE2I
VPS9D1-AS1
YWHAG
ZBTB16
ZCCHC12
ZMYND8
|
| Entrez ID |
10313 |
8405 |
| HPRD ID |
18516
|
16004
|
| Ensembl ID |
ENSG00000133318
|
ENSG00000121067
|
| Uniprot IDs |
O95197
|
O43791
|
| PDB IDs |
7BRU
|
2CR2
3HQH
3HQI
3HQL
3HQM
3HSV
3HTM
3HU6
3IVB
3IVQ
3IVV
4EOZ
4HS2
4J8Z
4O1V
6F8F
6F8G
6I41
6I5P
6I68
6I7A
7D3D
7KLZ
7LIN
7LIO
7LIP
7LIQ
8DWS
8DWT
8DWU
8DWV
|
| GO Terms Enriched among Interactors |
|
|