| Name |
axin 2 |
transcription elongation factor A N-terminal and central domain containing |
| Structure |
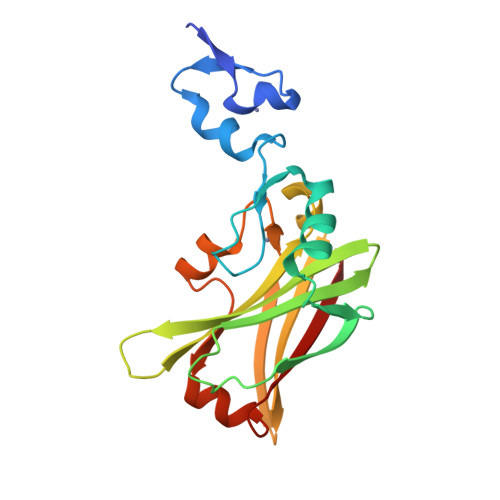
|
No pdb structure
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
None
|
| Diseases |
|
None
|
| GWAS |
|
No GWAS studies recorded
|
| Interacting Genes |
56 interacting genes:
AMOT
ANKRD6
AP1M1
APC
APC2
BARD1
BTRC
BYSL
C2orf68
CAPN7
CCDC112
CHIC2
CSNK1E
CTAG2
CTNNB1
CWF19L2
ELOA
FAM161A
FBP1
FBP2
FXR1
FXR2
GSK3B
HEMGN
HOXB5
KCNK12
KIFC3
KRTAP10-10
KRTAP10-8
KRTAP4-5
KRTAP9-2
LNX1
MCRS1
PPP2R5A
PRDX1
PSMB2
RANGAP1
SCNM1
SIAH1
SMAD1
SMAD3
SMAD7
SNCA
STX11
TCEA2
TCEANC
TGFB1
TOP3B
TSGA10
TXLNA
TXLNB
ZNF124
ZNF189
ZNF417
ZNF587
ZNF648
|
140 interacting genes:
ADARB1
AKAP9
ANAPC11
APP
ATXN1
AXIN2
BICD2
BRCA1
C1QTNF2
C8orf34
CARD9
CBY2
CCDC102B
CCDC136
CCDC138
CCDC33
CCDC85B
CCNC
CDR2L
CEP70
CEP76
CRACR2B
CREB3L3
CSRNP1
CTAG1A
CTAG1B
CYSRT1
DAB1
DISC1
DMAP1
DTX3
DVL3
DYNLT1
EVI5
FAM217B
FAM9B
FANCG
FBXO25
FCHO1
FSD2
GOLGA2
GOLGA6L9
HES7
HIP1
HMBOX1
HOMEZ
HOOK2
IGF1
IKBKG
IKZF2
IKZF3
KATNBL1
KCTD6
KCTD7
KIAA1958
KIF16B
KIFC3
KLHL12
KRT31
KRT35
KRT40
KRTAP1-1
KRTAP10-7
KRTAP10-8
KRTAP12-3
KRTAP4-2
KRTAP5-9
L3MBTL3
LDOC1
LHX2
LHX3
LIN7B
LMNA
LSM14B
LURAP1
MAGEA11
MCC
MCIDAS
MDFI
MEOX1
MEOX2
MID1
MID2
MTUS2
MYF5
MYLIP
NAB2
NECAB1
NEDD4
NINL
OBI1
OOEP
OSBPL3
PICK1
PLEKHF2
PNMA1
PNMA5
PXN
RAD54B
RNF41
RUBCN
SIPA1L2
SORBS3
SSNA1
STRN3
STX1A
STX4
TARBP2
TCP10L
THAP1
TLE5
TMCO2
TNIP1
TNIP3
TRAF2
TRIM23
TRIM27
TRIM32
TRIM35
TRIM37
TRIM41
TRIM54
UBXN11
USH1G
USHBP1
VAC14
VBP1
VPS52
YWHAE
ZBTB14
ZBTB39
ZBTB9
ZFYVE26
ZKSCAN7
ZNF276
ZNF655
ZNF670
ZNF688
ZNF837
ZRANB1
|
| Entrez ID |
8313 |
170082 |
| HPRD ID |
04935
|
19501
|
| Ensembl ID |
ENSG00000168646
|
ENSG00000176896
|
| Uniprot IDs |
B7ZKL5
E7ES00
Q9Y2T1
|
A8K5F6
B4DG85
Q8N8B7
|
| PDB IDs |
8HEO
|
Not available
|
| GO Terms Enriched among Interactors |
|
|