| Name |
troponin T3, fast skeletal type |
troponin I2, fast skeletal type |
| Structure |
No pdb structure
|
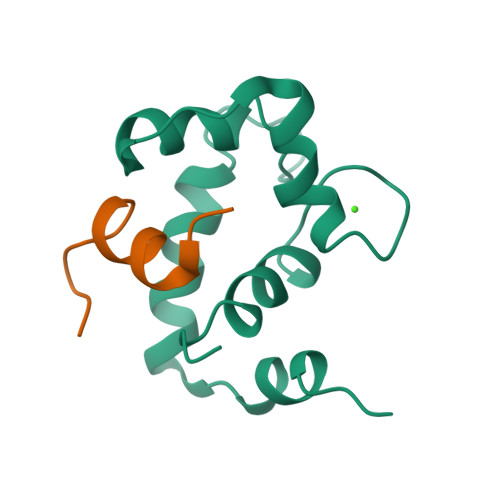
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
|
| Diseases |
|
|
| GWAS |
- Appendicular lean mass ( 33097823
)
- Celiac disease ( 23936387
)
- Diastolic blood pressure ( 27618452
)
- Diastolic blood pressure (cigarette smoking interaction) ( 29455858
)
- Diastolic blood pressure x smoking status (current vs non-current) interaction (2df test) ( 29455858
)
- Diastolic blood pressure x smoking status (ever vs never) interaction (2df test) ( 29455858
)
- Hypertension ( 30487518
)
- Mammographic density (dense area) ( 25342443
)
- Mean arterial pressure ( 27618448
30487518
)
- Pulse pressure x alcohol consumption interaction (2df test) ( 29912962
)
- Systolic blood pressure ( 27618452
)
- Systolic blood pressure (cigarette smoking interaction) ( 29455858
)
- Systolic blood pressure x smoking status (current vs non-current) interaction (2df test) ( 29455858
)
- Systolic blood pressure x smoking status (ever vs never) interaction (2df test) ( 29455858
)
|
- Celiac disease ( 23936387
)
- Diastolic blood pressure x smoking status (current vs non-current) interaction (2df test) ( 29455858
)
- Diastolic blood pressure x smoking status (ever vs never) interaction (2df test) ( 29455858
)
- Inflammatory bowel disease ( 23128233
)
- Logical memory (delayed recall) in mild cognitive impairment ( 29274321
)
- Systolic blood pressure x smoking status (current vs non-current) interaction (2df test) ( 29455858
)
- Systolic blood pressure x smoking status (ever vs never) interaction (2df test) ( 29455858
)
|
| Interacting Genes |
10 interacting genes:
CLIP4
HAP1
NLGN3
NUDT3
SNUPN
TNNI2
TRIM63
TSG101
TULP3
WASL
|
21 interacting genes:
ACTB
AIRIM
CALM1
CTNNA3
DCAF11
DRC8
ESRRA
ESRRG
HNF4G
MNS1
NME7
PKD2L1
PSMC5
RORB
TNNC1
TNNC2
TNNT1
TNNT3
TRIM55
TRIM63
UPF2
|
| Entrez ID |
7140 |
7136 |
| HPRD ID |
02822
|
01843
|
| Ensembl ID |
ENSG00000130595
|
ENSG00000130598
|
| Uniprot IDs |
A0A286YFB1
A0A9L9PY19
H9KVA2
P45378
|
P48788
|
| PDB IDs |
Not available
|
2MKP
7KAA
|
| GO Terms Enriched among Interactors |
- Vesicle Localization
- Establishment Of Vesicle Localization
- Nucleus
- Positive Regulation Of Cellular Component Biogenesis
- Regulation Of Epidermal Growth Factor Receptor Signaling Pathway
- Establishment Of Organelle Localization
- Regulation Of ERBB Signaling Pathway
- Establishment Of Localization In Cell
- Positive Regulation Of Ubiquitin-dependent Endocytosis
- Positive Regulation Of Viral Budding Via Host ESCRT Complex
- Intraciliary Transport Particle A Binding
- Bronchus Morphogenesis
- Negative Regulation Of Membrane Tubulation
- Vesicle Cytoskeletal Trafficking
- Positive Regulation Of Clathrin-dependent Endocytosis
- Brain-derived Neurotrophic Factor Binding
- Regulation Of Inositol 1,4,5-trisphosphate-sensitive Calcium-release Channel Activity
- Positive Regulation Of Neurotrophin Production
- Positive Regulation Of Inositol 1,4,5-trisphosphate-sensitive Calcium-release Channel Activity
- SnRNA Import Into Nucleus
- 5'-(N(7)-methyl 5'-triphosphoguanosine)-[mRNA] Diphosphatase Activity
- Inositol Diphosphate Tetrakisphosphate Diphosphatase Activity
- Diphosphoinositol Polyphosphate Metabolic Process
- Inositol Diphosphate Pentakisphosphate Diphosphatase Activity
- Inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase Activity
- Diphosphoinositol Polyphosphate Catabolic Process
- Bis(5'-adenosyl)-hexaphosphatase Activity
- Diadenosine Hexaphosphate Catabolic Process
- Inositol Phosphate Catabolic Process
- Diadenosine Pentaphosphate Catabolic Process
- Adenosine 5'-(hexahydrogen Pentaphosphate) Catabolic Process
- Diphosphoinositol-polyphosphate Diphosphatase Activity
- Endopolyphosphatase Activity
- Inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate Diphosphatase Activity
- Rhythmic Synaptic Transmission
- Symmetric, GABA-ergic, Inhibitory Synapse
- Response To Electrical Stimulus Involved In Regulation Of Muscle Adaptation
- Troponin T Binding
- Exosomal Secretion
- Organelle Localization
- Smoothened Signaling Pathway Involved In Dorsal/ventral Neural Tube Patterning
- Positive Regulation Of Plasma Membrane Bounded Cell Projection Assembly
- Actin Cap
- RNA Import Into Nucleus
- Bis(5'-adenosyl)-pentaphosphatase Activity
- Diadenosine Polyphosphate Catabolic Process
- Asymmetric, Glutamatergic, Excitatory Synapse
- Positive Regulation Of Cellular Component Organization
- Hypothalamus Cell Differentiation
- Skeletal Muscle Atrophy
|
|