| Name |
cortactin |
ribosomal protein L37 |
| Structure |
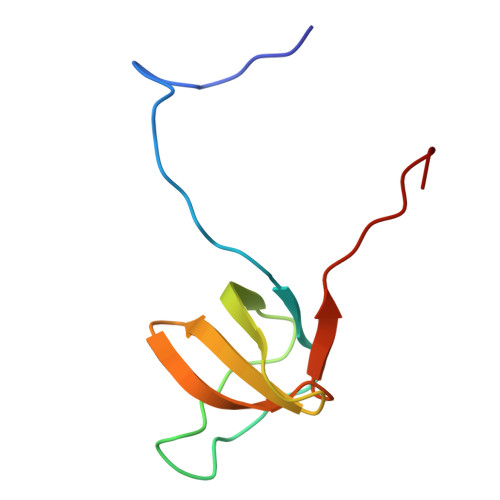
|
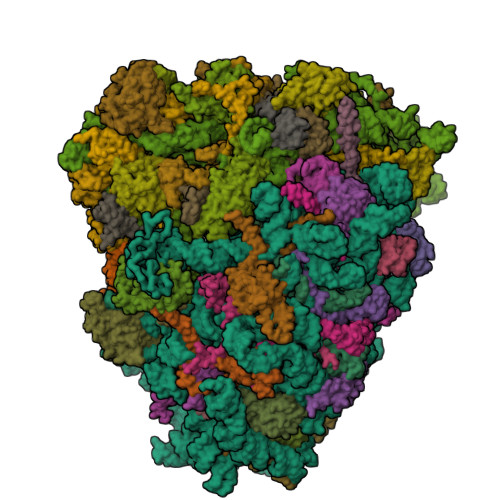
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
|
| Drugs |
None
|
|
| GWAS |
No GWAS studies recorded
|
No GWAS studies recorded
|
| Interacting Genes |
58 interacting genes:
ACD
ACTA1
ACTG1
ACTR3
ANKZF1
ARHGAP17
ARHGAP8
ATM
CASP3
CD2AP
CHD3
CPSF7
CTNND1
CTNND2
CTTNBP2
DNM1
DNM2
EGFR
FER
FGD1
GRB2
GRIA2
GTSE1
HAX1
HIP1R
KCNA2
KEAP1
MET
MYLK
NHSL2
NXF1
PRPF6
RBM15
RPL23A
RPL37
RPL9
RSPH1
SDC3
SF1
SF3B4
SFPQ
SH3BP2
SHANK2
SHANK3
SIRT1
SMURF1
SPRR2A
SRC
SYK
TINF2
TJP1
TNK2
TRIM15
USP7
WASL
WIPF1
YTHDF1
YTHDF3
|
9 interacting genes:
APP
CTTN
MYO1D
POLR1B
SQSTM1
SRPK2
TCF3
UBC
WIF1
|
| Entrez ID |
2017 |
6167 |
| HPRD ID |
01268
|
16047
|
| Ensembl ID |
ENSG00000085733
|
ENSG00000145592
|
| Uniprot IDs |
Q14247
Q53HG7
|
P61927
|
| PDB IDs |
1X69
2D1X
|
4UG0
4V6X
5AJ0
5LKS
5T2C
6IP5
6IP6
6IP8
6LQM
6LSR
6LSS
6LU8
6OLE
6OLF
6OLG
6OLI
6OLZ
6OM0
6OM7
6QZP
6W6L
6XA1
6Y0G
6Y2L
6Y57
6Y6X
6Z6L
6Z6M
6Z6N
6ZM7
6ZME
6ZMI
6ZMO
7BHP
7F5S
7OW7
7XNX
7XNY
8A3D
8FKP
8FKQ
8FKR
8FKS
8FKT
8FKU
8FKV
8FKW
8FKX
8FKY
8FKZ
8FL2
8FL3
8FL4
8FL6
8FL7
8FL9
8FLA
8FLB
8FLC
8FLD
8FLE
8FLF
8G5Y
8G5Z
8G60
8G61
8G6J
8GLP
8IDT
8IDY
8IE3
8IFD
8IFE
8INE
8INF
8INK
8IPD
8IPX
8IPY
8IR1
8IR3
8JDJ
8JDK
8JDL
8JDM
8K2C
8OHD
8OJ0
8OJ5
8OJ8
8QFD
8QOI
8QYX
8RL2
8UKB
8XSX
8XSY
8XSZ
8Y0W
8Y0X
8YOO
8YOP
9C3H
9G8M
9GMO
|
| GO Terms Enriched among Interactors |
|
|