| Name |
MAPK activated protein kinase 2 |
actin related protein 2/3 complex subunit 5 |
| Structure |
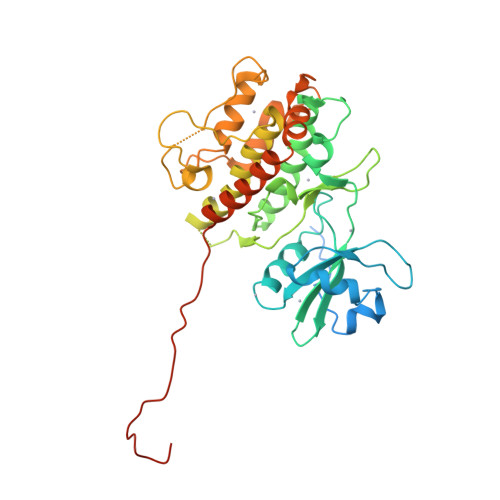
|
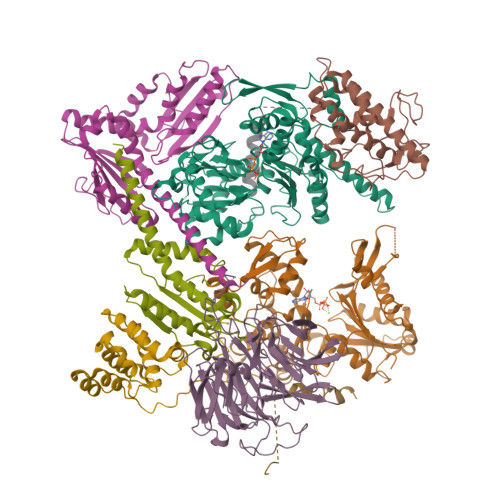
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
|
| Drugs |
- Staurosporine
- 3-{[(1R)-1-phenylethyl]amino}-4-(pyridin-4-ylamino)cyclobut-3-ene-1,2-dione
- (4R)-N-[4-({[2-(DIMETHYLAMINO)ETHYL]AMINO}CARBONYL)-1,3-THIAZOL-2-YL]-4-METHYL-1-OXO-2,3,4,9-TETRAHYDRO-1H-BETA-CARBOLINE-6-CARBOXAMIDE
- (10R)-10-methyl-3-(6-methylpyridin-3-yl)-9,10,11,12-tetrahydro-8H-[1,4]diazepino[5',6':4,5]thieno[3,2-f]quinolin-8-one
- (3R)-3-(aminomethyl)-9-methoxy-1,2,3,4-tetrahydro-5H-[1]benzothieno[3,2-e][1,4]diazepin-5-one
- 2-[2-(2-FLUOROPHENYL)PYRIDIN-4-YL]-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE
- 2-(2-QUINOLIN-3-YLPYRIDIN-4-YL)-1,5,6,7-TETRAHYDRO-4H-PYRROLO[3,2-C]PYRIDIN-4-ONE
|
|
| GWAS |
- Inflammatory bowel disease ( 23128233
)
- Itch intensity from mosquito bite ( 28199695
)
- Mean corpuscular hemoglobin ( 32888494
)
|
No GWAS studies recorded
|
| Interacting Genes |
42 interacting genes:
AGO2
AKT1
ALOX5
ARPC5
ATF1
BAG2
BCL2L1
BECN1
CDC25B
CEP131
CREB1
DDX5
DES
EEF2K
ETV1
HNRNPA0
HSF1
HSPB1
LIMK1
LSP1
MAP2K6
MAPK14
MAPK3
MDM2
PABPC1
PARN
PHC2
PIAS2
RCSD1
SHC1
SRF
STAT3
TCF3
TH
TRIM28
TRIM29
TSC2
UBE2J1
WASF1
YWHAZ
ZFP36
ZFP36L1
|
6 interacting genes:
ACTR2
ACTR3
ARPC1B
ARPC4
MAPKAPK2
NR3C1
|
| Entrez ID |
9261 |
10092 |
| HPRD ID |
11882
|
11969
|
| Ensembl ID |
ENSG00000162889
|
ENSG00000162704
|
| Uniprot IDs |
P49137
|
O15511
|
| PDB IDs |
1KWP
1NXK
1NY3
2JBO
2JBP
2OKR
2ONL
2OZA
2P3G
2PZY
3A2C
3FPM
3FYJ
3FYK
3GOK
3KA0
3KC3
3KGA
3M2W
3M42
3R2B
3R2Y
3R30
3WI6
4TYH
6T8X
6TCA
7NRY
8XU4
8XX1
|
6UHC
6YW7
|
| GO Terms Enriched among Interactors |
|
|