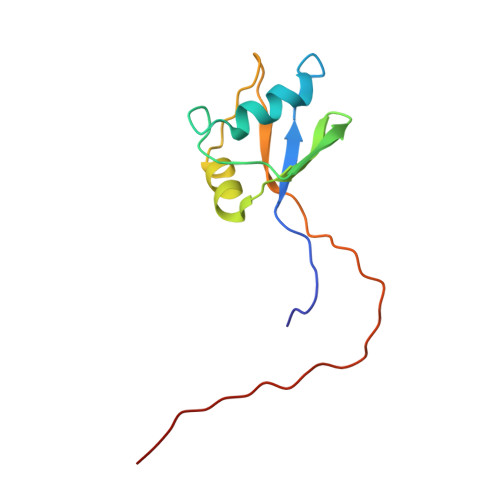
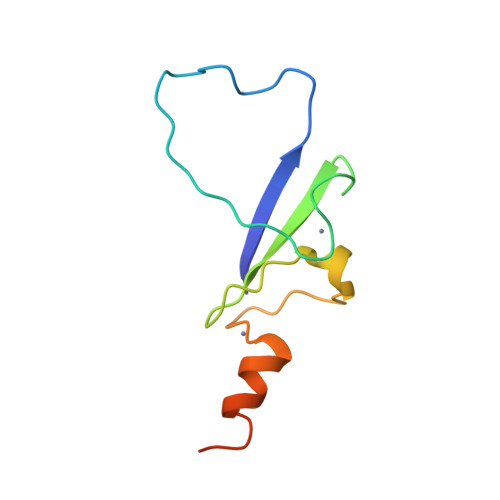
| Name |
poly(ADP-ribose) polymerase family member 10 |
H3.3 histone A |
| Structure |
|
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
|
| GWAS |
|
No GWAS studies recorded
|
| Interacting Genes |
14 interacting genes:
AURKA
DTX2
H2AC20
H2BC21
H3-3A
H3C1
H4C1
MYC
PARP14
PCNA
PLK1
RAN
RNF26
UBC
|
44 interacting genes:
ASF1A
ASF1B
BPTF
BRD7
CARM1
CBX2
CBX4
CBX6
CBX7
CBX8
CEBPA
CHD1
CREBBP
DCAF1
EP300
ESR1
HMGB2
HPF1
KAT2B
KMT2A
MAPK9
NEDD4
NOS3
PARP10
PARP2
PELP1
PIM1
PPM1D
PPP1CA
PPP1CB
SET
SETD7
SMARCA1
SMARCA4
SMARCA5
SS18
SSRP1
SUPT16H
SUV39H1
TIAM1
TLE2
UBN1
UHRF1
ZMYND11
|
| Entrez ID |
84875 |
3020 |
| HPRD ID |
15100
|
03082
|
| Ensembl ID |
ENSG00000178685
|
ENSG00000163041
|
| Uniprot IDs |
B4E0C4
E9PK67
E9PNI7
E9PPE7
Q53GL7
|
B2R4P9
P84243
|
| PDB IDs |
2DHX
3HKV
5LX6
6FXI
|
2L43
3ASK
3ASL
3AV2
3JVK
3MUK
3MUL
3QL9
3QLA
3QLC
3WTP
4GNE
4GNF
4GNG
4GU0
4GUR
4GUS
4GY5
4H9N
4H9O
4H9P
4H9Q
4H9R
4H9S
4HGA
4L58
4N4I
4O62
4QQ4
4TMP
4U7T
4W5A
5B32
5B33
5BNV
5BNX
5DWQ
5DX0
5JA4
5JJY
5JLB
5KDM
5X7X
6A5L
6A5O
6A5P
6A5R
6A5T
6A5U
6HGT
6INQ
6IR9
6J4W
6J4X
6J4Y
6J4Z
6J50
6J51
6J9J
6PZV
6QXZ
6R0C
6RNY
6U04
7A08
7CIZ
7CJ0
7CWH
7OKP
7V1L
7V1M
7VCQ
7W5M
7WBV
7WBW
7WBX
7XSE
7XSX
7XSZ
7XT7
7XTD
7XTI
8JH2
8JH3
8JH4
8KB5
9B3P
9EOZ
9J0N
9J0O
9J0P
|
| GO Terms Enriched among Interactors |
|
|