| Name |
transmembrane serine protease 2 |
C-type lectin domain containing 7A |
| Structure |
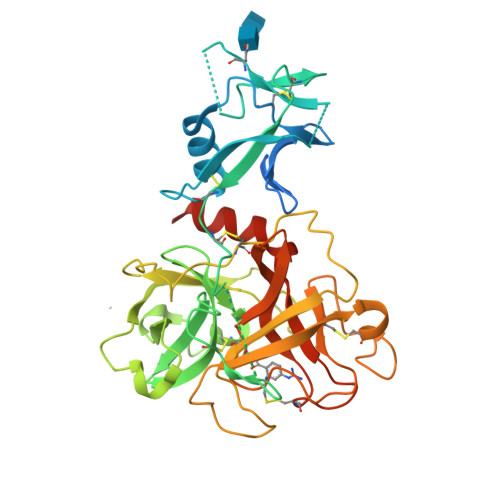
|
No pdb structure
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
|
| Drugs |
|
None
|
| Diseases |
|
|
| GWAS |
- Amyotrophic lateral sclerosis (sporadic) ( 24529757
)
- Liver enzyme levels (gamma-glutamyl transferase) ( 33972514
)
- Prostate cancer ( 25217961
)
|
No GWAS studies recorded
|
| Interacting Genes |
60 interacting genes:
ACE2
ADIPOQ
ANKRD46
AQP1
BMP10
BNIP2
BNIP3
BRICD5
C1QL4
C2CD2L
C3orf52
CFHR5
CLEC7A
CLK1
CLN6
CMTM7
CNIH2
CNIH3
CTXN3
CXCL9
CYBC1
DEFB103A
DEFB103B
EDDM3B
ERG28
FAM3C
FAXDC2
IGFBP5
MOSPD3
NINJ2
PGAP2
PLLP
PLN
PLP1
PLP2
PTCH1
PTTG1IP
SCD
SEC22A
SELENOK
SFTPC
SLC35A1
SMIM1
STX7
STX8
TMEM11
TMEM120B
TMEM128
TMEM218
TMEM222
TMEM229B
TMEM243
TMEM60
TMEM79
TMEM86A
TMEM86B
TNF
UPK2
VAMP5
ZFPL1
|
48 interacting genes:
ADAM33
AIP
ARL13B
ASGR2
BNIP3
BNIP3L
C14orf180
CALM3
CD53
CD74
CFTR
CTXN3
CXCL9
FAM3A
FCGR1A
FNDC9
GJB1
HSPD1
IFNGR2
IL1RL1
JAGN1
KLRC1
KTN1
MTIF3
NEMP1
NINJ2
NSG2
OCLN
OLFM4
OPRM1
PTTG1IP
RANBP9
RHOU
SAR1A
SEC22A
SGCB
SMIM3
SPINT1
SUSD3
SYNE4
TEX29
TLCD4
TMEM79
TMPRSS2
TMPRSS4
UPK2
VSIR
ZFPL1
|
| Entrez ID |
7113 |
64581 |
| HPRD ID |
03637
|
08396
|
| Ensembl ID |
ENSG00000184012
|
ENSG00000172243
|
| Uniprot IDs |
A0A7I2V474
O15393
|
A0A0S2Z5Q1
A0A2Z5YRT6
A0A2Z5YRU4
Q68D78
Q9BXN2
|
| PDB IDs |
7MEQ
7XYD
7Y0E
7Y0F
8HD8
8JHZ
8JI0
8S0L
8S0M
8S0N
8V04
8V1F
8VGT
8Y1D
8Y1E
8Y7X
8Y7Y
8Y87
8Y88
8Y89
8Y8A
8Y8B
8YOY
8YQQ
9IZN
|
Not available
|
| GO Terms Enriched among Interactors |
|
|