| Name |
ATPase H+ transporting V0 subunit b |
Fas cell surface death receptor |
| Structure |
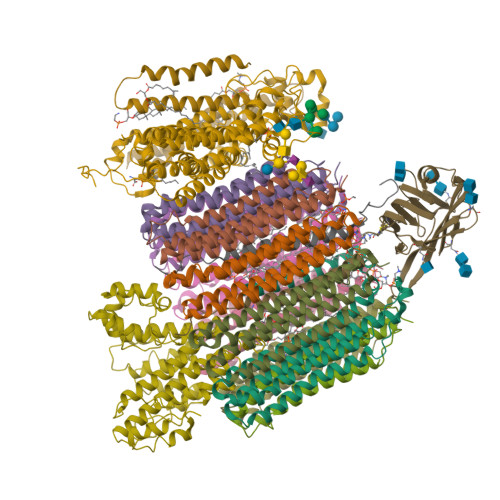
|
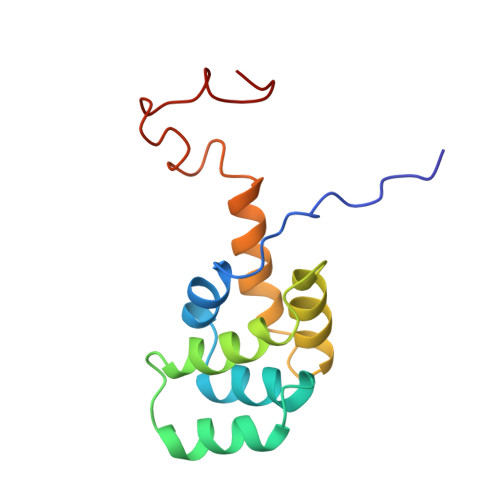
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
|
| Drugs |
|
None
|
| Diseases |
None
|
|
| GWAS |
- Autism spectrum disorder or schizophrenia ( 28540026
)
|
|
| Interacting Genes |
28 interacting genes:
ADGRG3
ADRB2
APOD
COL4A5
EMP1
FAM210B
FAS
GJA8
GPR42
GPR61
INSIG2
IRAK3
KLRC1
MFF
MIP
MS4A3
MTNR1B
NRM
SLC10A1
SLC2A5
SNTA1
SPACA1
STOM
TM4SF19
TMEM242
VMA21
YWHAE
YWHAG
|
50 interacting genes:
ANK3
APAF1
APOA5
ATP6V0B
BAG6
BTK
CALM1
CASP8
CASP8AP2
CCND3
CD47
CRMP1
DAXX
DNAJC5
EEF1A1
EGFR
ERG28
EZR
FADD
FAF1
FAF2
FAIM2
FASLG
FBF1
FEM1B
FYN
GADD45A
HIPK3
HNRNPC
LCK
LRIF1
MBD4
MET
PDCD6
PRKCA
PTPN13
PTPN6
RAP1A
RIPK1
SUMO1
TCAP
TMX1
TNFSF13
TRADD
TRAF3
TRIP6
UBA7
UBAC1
UBE2I
UBQLN1
|
| Entrez ID |
533 |
355 |
| HPRD ID |
04759
|
00609
|
| Ensembl ID |
ENSG00000117410
|
ENSG00000026103
|
| Uniprot IDs |
E9PNL3
Q99437
|
A0A8Q3SIR6
K9J972
P25445
Q59FU8
|
| PDB IDs |
6WLW
6WM2
6WM3
6WM4
7U4T
7UNF
|
1DDF
2NA7
3EWT
3EZQ
3THM
3TJE
|
| GO Terms Enriched among Interactors |
|
|