| Name |
cyclin D3 |
activating transcription factor 5 |
| Structure |
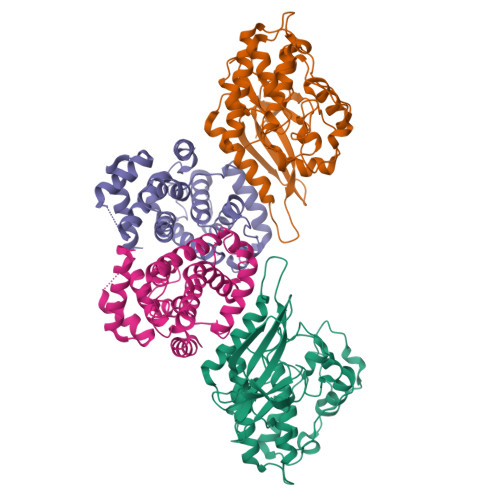
|
No pdb structure
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
|
| Drugs |
None
|
|
| GWAS |
|
No GWAS studies recorded
|
| Interacting Genes |
57 interacting genes:
AKAP8
APP
AREG
ATF5
ATP5F1C
C14orf119
CASP2
CCDC13
CDK11B
CDK14
CDK16
CDK18
CDK3
CDK4
CDK5
CDK6
CDKN1A
CDKN1B
CDKN2D
CRABP2
DMRTB1
DMTF1
DTNBP1
EFEMP2
EIF3K
ETV1
FAS
FHL3
GC
HGS
KIFC3
LSM3
MAF1
MAPK4
MAPK6
MCM10
MNS1
MYB
NCOA2
NME1
NPDC1
PCNA
POLD1
POLR3GL
PSMC1
QKI
RARA
RB1
RBL2
RUNX1
SH3GLB2
SIL1
TEX11
TFIP11
TSG101
USP10
VDR
|
18 interacting genes:
ATXN1
CCDC6
CCND3
CDC34
CEBPA
CEBPE
CEBPG
DISC1
EIF2B1
EP300
FBXO7
GABBR1
GABBR2
GIT2
GPS2
KEAP1
SYCE1L
TTR
|
| Entrez ID |
896 |
22809 |
| HPRD ID |
00452
|
05906
|
| Ensembl ID |
ENSG00000112576
|
ENSG00000169136
|
| Uniprot IDs |
B3KP19
P30281
Q5T8J1
|
Q9Y2D1
|
| PDB IDs |
3G33
7SJ3
|
Not available
|
| GO Terms Enriched among Interactors |
|
|