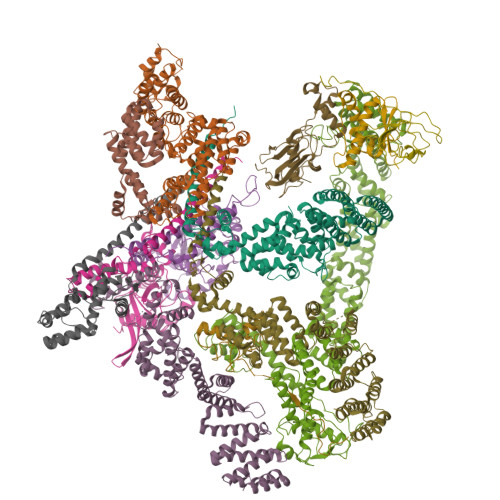
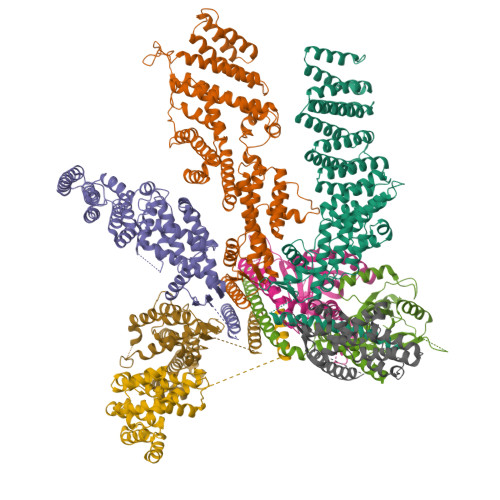
| Name |
COP9 signalosome subunit 7B |
eukaryotic translation initiation factor 3 subunit E |
| Structure |
|
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
|
| GWAS |
|
|
| Interacting Genes |
6 interacting genes:
APP
BEX5
EIF3E
NMI
PMF1
UBC
|
37 interacting genes:
ANKHD1
ATM
COPS6
COPS7A
COPS7B
COPS8
DDX24
EIF3C
EIF3L
EIF4ENIF1
EPAS1
EPN2
GPAA1
GPBP1L1
HAP1
IFIT1
ISCA2
KPRP
LINC01554
MAPK1IP1L
MIIP
MRNIP
MSI2
NDRG1
NSF
OGT
PGCKA1
PRPF31
PRRC2A
RPSA
RUNDC3A
SHBG
SMAD9
TRIM27
TRIM55
TRIM63
ZNF48
|
| Entrez ID |
64708 |
3646 |
| HPRD ID |
09889
|
03734
|
| Ensembl ID |
ENSG00000144524
|
ENSG00000104408
|
| Uniprot IDs |
A0A087X1P5
J3KQ34
J3KQ41
Q6ZTQ7
Q9H9Q2
|
P60228
|
| PDB IDs |
6R6H
6R7F
6R7H
6R7I
6R7N
8H38
8H3A
8H3F
|
3J8B
3J8C
6FEC
6YBD
6ZMW
6ZON
6ZP4
6ZVJ
7A09
7QP6
7QP7
8OZ0
8PJ1
8PJ2
8PJ3
8PJ4
8PJ5
8PJ6
8PPL
8RG0
8XXN
9BLN
|
| GO Terms Enriched among Interactors |
|
|