| Name |
actinin alpha 2 |
angiogenin |
| Structure |
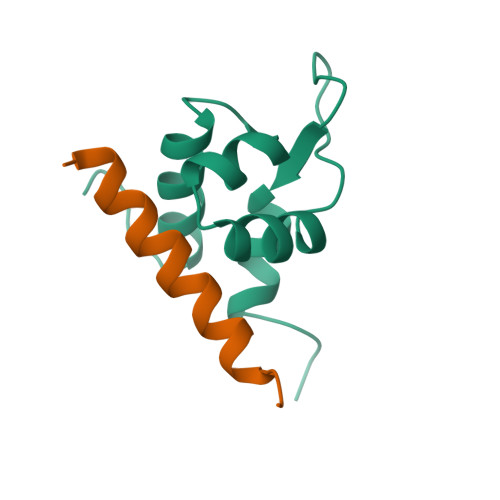
|
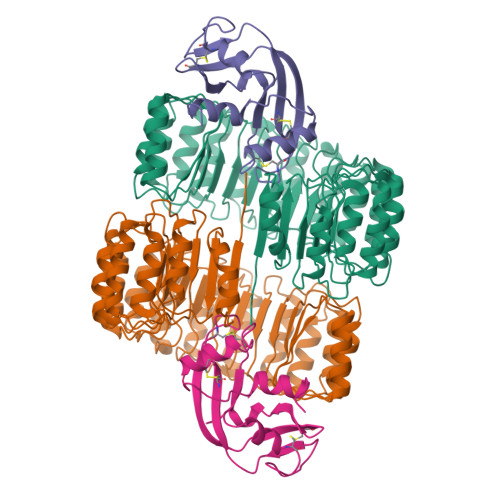
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
|
| Diseases |
None
|
|
| GWAS |
|
|
| Interacting Genes |
110 interacting genes:
ACTN3
ADAM12
ADORA2A
AKTIP
ANG
ANGPTL7
ARX
ASH2L
ATP5MC1
ATXN2
ATXN7
BAD
BRMS1L
CACNA1C
CAMK2A
CAMK2D
CAMK2G
CAPN1
CCDC187
CD27
CIMAP1B
CLEC4D
CNNM3
COIL
CRABP2
CRADD
DISC1
DLG1
DLG4
DUX1
DYNLT2B
EPS8L1
ERBIN
FAM50B
FBXL22
FXR1
GATA3
GOLGA7
GRIN1
GRIN2B
GSTT1
H4C9
HSPB1
HTR1B
ITGB3BP
KAT2B
KATNAL2
KCNA4
KCNA5
KCNN2
LDB3
LRP12
LRRC7
MAST2
MED14
MICALL2
MOS
MRPL10
MYBPC2
MYOT
MYOZ1
MYOZ2
MYOZ3
MYPN
NCAPH2
NCOA2
NCOR1
NKAPD1
NOS3
NR1I2
NRIP1
NTAQ1
PALLD
PDLIM1
PDLIM3
PKD2
PPP1CB
PPP1R9B
PSMA1
QARS1
RACK1
RAVER1
RPL35
RPP14
RTP5
SAXO1
SELE
SHANK3
SMARCA2
SNAI1
SNAPIN
SNW1
SP100
SPA17
SRP9
SSX2IP
ST7
SYNPO2
SYNPO2L
TNN
TOLLIP
TSC1
TSC2
TTN
TULP3
USP2
UTRN
ZC2HC1C
ZNF446
ZNRD2
|
8 interacting genes:
ABCG8
ACTC1
ACTN2
ATP6AP1
CRIPTO
PTEN
RNH1
TNFSF8
|
| Entrez ID |
88 |
283 |
| HPRD ID |
00019
|
00105
|
| Ensembl ID |
ENSG00000077522
|
ENSG00000214274
|
| Uniprot IDs |
P35609
|
P03950
W0UV28
|
| PDB IDs |
1H8B
1HCI
1QUU
4D1E
5A36
5A37
5A38
5A4B
6SWT
6TS3
7A8T
7A8U
7B55
7B56
7B57
|
1A4Y
1ANG
1AWZ
1B1E
1B1I
1B1J
1GV7
1H0D
1H52
1H53
1HBY
1K58
1K59
1K5A
1K5B
1UN3
1UN4
1UN5
2ANG
4AHD
4AHE
4AHF
4AHG
4AHH
4AHI
4AHJ
4AHK
4AHL
4AHM
4AHN
4AOH
4B36
5EOP
5EPZ
5EQO
5M9A
5M9C
5M9G
5M9J
5M9M
5M9P
5M9Q
5M9R
5M9S
5M9T
5M9V
7NPM
7PNJ
7PNP
7PNR
8AF0
9BDL
9BDN
9BDP
|
| GO Terms Enriched among Interactors |
|
|