| Name |
centromere protein S |
homeobox and leucine zipper encoding |
| Structure |
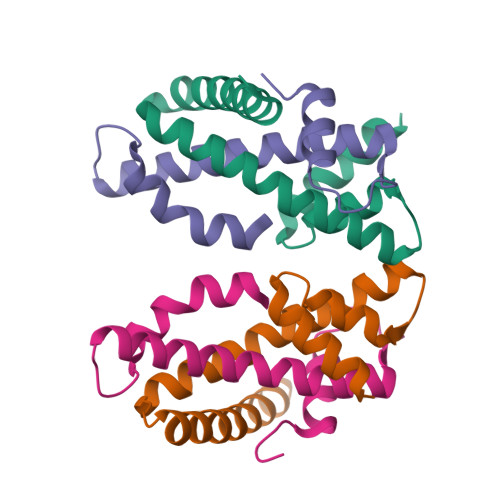
|
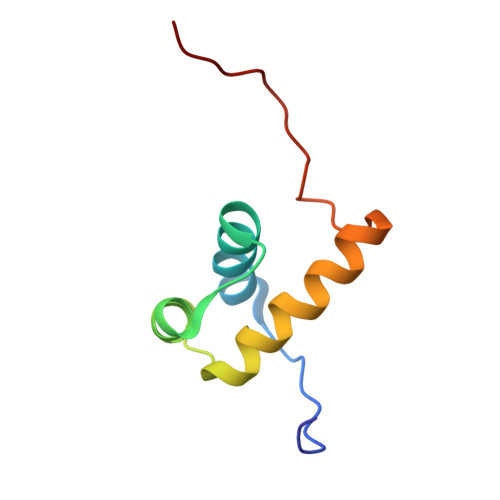
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
None
|
| GWAS |
No GWAS studies recorded
|
- Height ( 31562340
)
- High light scatter reticulocyte count ( 32888494
)
- High light scatter reticulocyte percentage of red cells ( 32888494
)
- Hip circumference adjusted for BMI ( 34021172
)
- Mean platelet volume ( 32888494
)
- Pulse pressure ( 27841878
)
- Reticulocyte count ( 32888494
)
- Serum alkaline phosphatase levels ( 33547301
)
- Systolic blood pressure ( 27841878
)
|
| Interacting Genes |
5 interacting genes:
APP
CENPM
CENPT
CENPX
HOMEZ
|
114 interacting genes:
ACTR10
AEN
ANKRD11
ANKRD2
ARHGAP9
ASB6
AXIN1
BIVM
BYSL
C2orf42
CBX8
CCDC13
CCDC187
CCHCR1
CCNK
CDC7
CENPS
CENPS-CORT
CFAP206
CTNNA3
DEF6
DGCR6
DHX16
DNTTIP1
DRC4
EBF1
EIF4E2
ELOA
ENKD1
FAM90A1
FARS2
GFAP
GPANK1
GRB2
HOXC8
HSF2BP
IL16
INO80B
JAKMIP2
KIFC3
KPNA2
LENG1
LGALS4
LMO1
LMO3
LNX1
LRRC7
LSM4
MAD2L1BP
MAFG
MAPK1
MED18
MORF4L2
MOS
MRPL11
NCK1
NCK2
NEIL2
NEK6
NXF1
OSGIN1
PARVG
PHF1
PHF19
PIBF1
PIN1
PLEKHA7
POLDIP3
PRKAA1
PRKAA2
PRPF18
PSMA1
RASL10A
RBM39
RNF8
RPGR
RPL9
RPP25
RPS25
SCNM1
SDCBP
SEC23A
SH3RF2
SMARCB1
SMARCD1
SMYD1
SNRPB2
SUMO2
SUMO3
TARS2
TBC1D30
TCEA2
TCEANC
TEAD4
TLE5
TRAF4
TSGA10IP
TXK
USF1
VEZF1
VPS9D1
ZBTB25
ZBTB7A
ZMYND19
ZNF165
ZNF250
ZNF280C
ZNF35
ZNF417
ZNF580
ZNF629
ZNF688
ZNF76
ZSCAN5B
|
| Entrez ID |
378708 |
57594 |
| HPRD ID |
18797
|
12167
|
| Ensembl ID |
ENSG00000175279
|
ENSG00000290292
|
| Uniprot IDs |
Q8N2Z9
|
Q8IX15
|
| PDB IDs |
4DRA
4DRB
4E44
4E45
4NDY
4NE1
4NE3
4NE5
4NE6
7R5S
7XHN
7XHO
7YWX
|
2ECC
2YS9
|
| GO Terms Enriched among Interactors |
|
|