| Name |
centromere protein S |
centromere protein M |
| Structure |
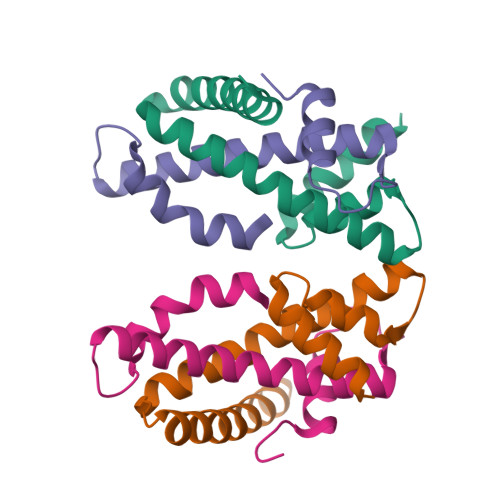
|
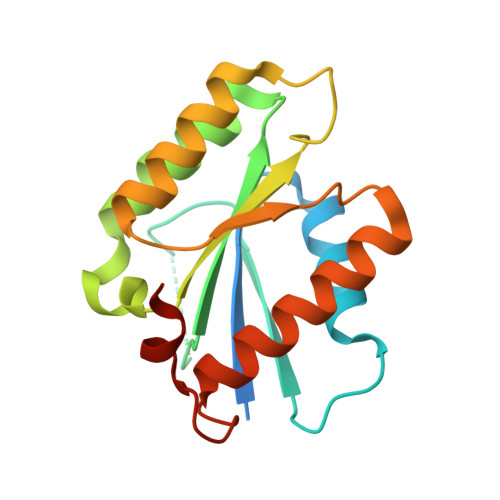
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
None
|
| Biological Process |
|
|
| Pathways |
|
|
| GWAS |
No GWAS studies recorded
|
|
| Interacting Genes |
5 interacting genes:
APP
CENPM
CENPT
CENPX
HOMEZ
|
2 interacting genes:
CENPS
H3C1
|
| Entrez ID |
378708 |
79019 |
| HPRD ID |
18797
|
12798
|
| Ensembl ID |
ENSG00000175279
|
ENSG00000100162
|
| Uniprot IDs |
Q8N2Z9
|
B1AHQ6
B1AHQ7
B1AHQ8
Q9NSP4
|
| PDB IDs |
4DRA
4DRB
4E44
4E45
4NDY
4NE1
4NE3
4NE5
4NE6
7R5S
7XHN
7XHO
7YWX
|
4P0T
4WAU
7PKN
7QOO
7R5S
7R5V
7XHN
7XHO
7YWX
7YYH
|
| GO Terms Enriched among Interactors |
|
None
|