| Name |
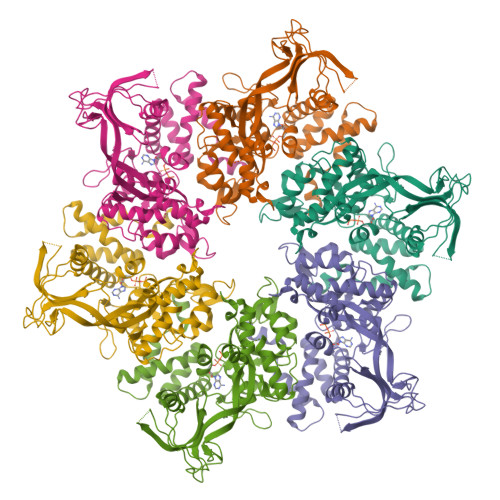
RuvB like AAA ATPase 1 |
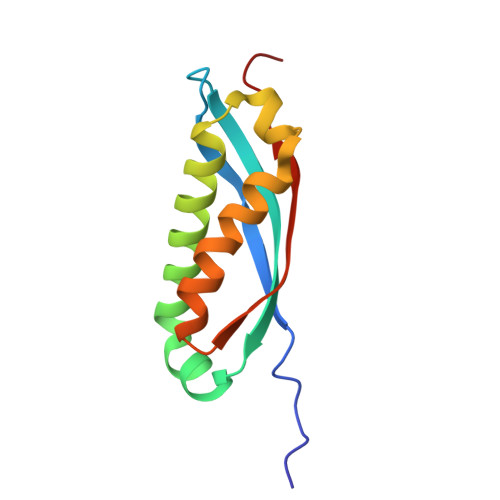
stomatin |
| Structure |
|
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
|
| Diseases |
None
|
|
| GWAS |
- Chronic obstructive pulmonary disease or high blood pressure (pleiotropy) ( 30940143
)
- Chronic obstructive pulmonary disease or resting heart rate (pleiotropy) ( 30940143
)
- Endometrial cancer ( 30093612
)
- Endometrial cancer (endometrioid histology) ( 30093612
)
- Gestational age at birth (maternal effect) ( 28877031
)
- Post bronchodilator FEV1 ( 26634245
)
- Post bronchodilator FEV1/FVC ratio ( 26634245
)
|
No GWAS studies recorded
|
| Interacting Genes |
20 interacting genes:
ACTL6A
CEP126
CTNNB1
DKC1
FBL
FOXP1
LZTR1
MRGBP
NEDD4
OFD1
OGT
PCBP3
PLG
RUVBL2
STOM
SUMO2
TAF9
TBP
TERT
TK1
|
81 interacting genes:
AIG1
APOA2
ASIC1
ASIC2
ASIC3
ATP1B3
ATP6V0B
ATP6V0C
BCL2L2
BNIP1
C2
C3orf52
CFHR5
CHST1
CLDN19
CLRN2
CMTM5
CNIH3
CTXN3
CYB5B
CYB5R3
DAGLA
DVL3
EMP1
ERG28
FAXDC2
FUNDC2
GIMAP1
GIMAP5
GPR25
GPR37L1
GRM2
HMOX2
KCNK1
KTN1
LANCL1
LNPEP
MGLL
MMP14
NKG7
ORMDL1
PEDS1-UBE2V1
PEMT
PGA4
PMP22
PTCH1
RPL13A
RPRM
RTP2
RUVBL1
RUVBL2
SCD
SELENOK
SERP2
SFT2D1
SFT2D2
SFXN1
SFXN5
SLC2A1
SLC35B4
SMCO4
STX6
TECR
THSD7B
TMEM109
TMEM120B
TMEM140
TMEM14B
TMEM203
TMEM208
TMEM254
TMEM60
TMEM86B
TMEM98
TMPO
TNF
TNFRSF10C
TSPO2
VAMP4
WFDC2
YIPF6
|
| Entrez ID |
8607 |
2040 |
| HPRD ID |
09143
|
00585
|
| Ensembl ID |
ENSG00000175792
|
ENSG00000148175
|
| Uniprot IDs |
A0A384MTR5
B3KRS7
E7ETR0
Q9Y265
|
F8VSL7
P27105
|
| PDB IDs |
2C9O
2XSZ
5OAF
6FO1
6HTS
6IGM
6K0R
6QI8
6QI9
7AHO
7OLE
7P6X
7ZI4
8QR1
8X15
8X19
8X1C
8XVG
8XVT
9C57
9C62
9EMA
9EMC
|
7WH3
|
| GO Terms Enriched among Interactors |
|
|