| Name |
nucleoporin 214 |
protein O-mannosyltransferase 1 |
| Structure |
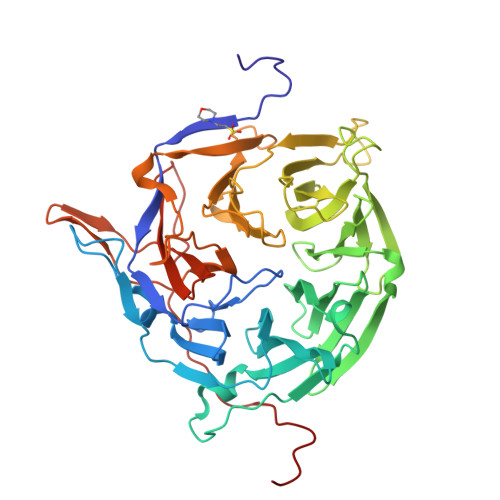
|
No pdb structure
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
|
| Diseases |
None
|
|
| GWAS |
- Aspartate aminotransferase levels ( 31219225
)
|
No GWAS studies recorded
|
| Interacting Genes |
19 interacting genes:
APC
CLEC4G
DDX19B
IPO5
NUP107
NUP42
NUP88
NXF1
NXF2
SMAD2
SMAD3
SMAD4
SUMO2
TNPO1
TSG101
XPO1
XPO5
XPOT
ZFP36
|
2 interacting genes:
MEOX2
POMT2
|
| Entrez ID |
8021 |
10585 |
| HPRD ID |
00258
|
06305
|
| Ensembl ID |
ENSG00000126883
|
ENSG00000130714
|
| Uniprot IDs |
B7ZAV2
P35658
|
A0A140VKE0
A0A7I2V2K0
A0A7I2V445
B4DTW4
Q5JT02
Q9Y6A1
|
| PDB IDs |
2OIT
3FHC
3FMO
3FMP
5DIS
7R5J
7R5K
|
Not available
|
| GO Terms Enriched among Interactors |
|
|