| Name |
dopamine beta-hydroxylase |
notch 2 N-terminal like A |
| Structure |
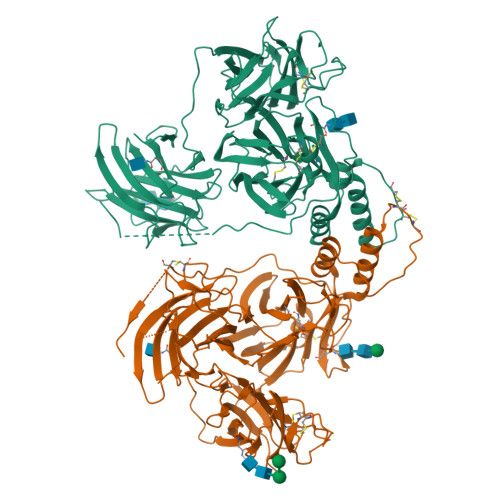
|
No pdb structure
|
| GO Annotations |
Cellular Component |
|
|
| Molecular Function |
|
|
| Biological Process |
|
|
| Pathways |
|
|
| Drugs |
|
None
|
| Diseases |
|
None
|
| GWAS |
|
No GWAS studies recorded
|
| Interacting Genes |
5 interacting genes:
CYSRT1
NBPF19
NOTCH2NLA
PRADC1
SNCA
|
361 interacting genes:
ACY3
ADAM12
ADAMTSL3
ADAMTSL4
ADAMTSL5
ADCK5
AGXT
ALDH16A1
ALDH3B1
ALPI
ALPP
ANKS1A
AQP1
AQP5
ARID3A
ASPSCR1
ATG9A
ATP5F1D
BCAM
BCL6B
BLCAP
BMP7
BRME1
C11orf87
CA6
CARHSP1
CATSPER1
CCDC26
CCDC93
CCER1
CD164
CDK5R1
CELF5
CERK
CHCHD3
CHDH
CHIC2
CHRD
CHRNG
CLDN2
CLEC18A
CNNM3
COL8A1
CRACR2A
CRCT1
CREB5
CRH
CRY2
CSF1
CST2
CST9L
CTNNBIP1
CTSG
CTSZ
CXCL16
CXCL5
CYP21A2
CYSRT1
DAGLB
DALRD3
DBH
DGCR6
DGKQ
DHRS1
DMRT3
DNAL4
DOCK2
DRD5
EFNA3
EIF4E2
ELANE
EMC7
EPHB6
ESR2
F10
FAAH
FADS2
FAM124B
FAM221A
FAM74A4
FAM90A1
FARS2
FASLG
FBXL18
FBXW5
FOXB1
FRS3
FTHL17
FZD10
GABRD
GARIN5B
GARIN6
GATA2
GEM
GFOD1
GIP
GLP1R
GLRX3
GLYAT
GNAI2
GNE
GNMT
GSTP1
GTF3C5
H2AC15
HAPLN2
HBA1
HBA2
HBZ
HCK
HHEX
HOXA1
HOXB9
HOXC8
HPCAL1
HSBP1
HSD3B7
HSPA12B
HSPBP1
HSPD1
ICAM4
IFI30
IGSF8
IL10
IL2RG
INO80B
INPP5D
INS
ITGB2
ITGB4
ITGB5
JOSD1
KCNS2
KCTD15
KIF1A
KLHL38
KLK8
KPRP
KRT20
KRT83
KRTAP10-1
KRTAP10-10
KRTAP10-11
KRTAP10-3
KRTAP10-5
KRTAP10-7
KRTAP10-8
KRTAP10-9
KRTAP12-1
KRTAP12-2
KRTAP12-3
KRTAP12-4
KRTAP13-2
KRTAP13-3
KRTAP13-4
KRTAP2-3
KRTAP2-4
KRTAP26-1
KRTAP3-1
KRTAP4-11
KRTAP4-12
KRTAP4-2
KRTAP4-4
KRTAP4-5
KRTAP4-7
KRTAP5-11
KRTAP5-3
KRTAP5-4
KRTAP5-6
KRTAP5-9
KRTAP9-2
KRTAP9-3
KRTAP9-4
KRTAP9-8
LCE1A
LCE1B
LCE1D
LCE1E
LCE1F
LCE2A
LCE2B
LCE2C
LCE2D
LCE3A
LCE3C
LCE3D
LCE3E
LCE4A
LCE5A
LIMS2
LIN7A
LINC00526
LINC00656
LMNTD2
LMO2
LNX1
LONRF3
LRCH4
LRFN4
LYVE1
MACO1
MAPKBP1
MARK4
MATN3
MELTF
MOS
MRGBP
MRPL40
MTA1
MVP
MXD3
MXI1
MYO15B
MYPOP
NAB2
NAXD
NECTIN2
NECTIN3
NEDD9
NEU2
NMU
NMUR2
NPBWR2
NPDC1
NPPB
NR1D2
NTM
NTN4
NUBP2
NUFIP2
OLFM2
OTX1
P2RX4
P2RX7
P2RY6
PCED1A
PCED1B
PCSK5
PDE9A
PGAP6
PGLS
PHLDA1
PID1
PIGS
PLEKHN1
PLPP2
PLSCR4
POLR2G
POM121L8P
POMGNT2
PRF1
PRKAA2
PRPF31
PRPS2
PRR19
PRR35
PSMA1
PSMG2
PTGER3
PTK7
PTPMT1
PTPN23
PVR
QPRT
R3HDM2
RAB3IL1
RAMP3
RASD1
RCHY1
RECK
RET
RGL2
RNF111
RPS19BP1
RPS28
RTN4RL1
SCNM1
SDCBP
SELENOM
SEMA3B
SEMA4C
SHANK3
SHFL
SLC22A23
SLC22A5
SLC23A1
SLC25A10
SLC25A6
SLC43A2
SLC5A5
SLC68A1
SLC6A20
SMARCD2
SMARCE1
SMCP
SMOC1
SNAI1
SPATA24
SPATA31J1
SPATA8
SPG7
SPINK2
SPPL2C
SPRY1
STAC
STK16
TAPBPL
TBC1D10C
TBC1D16
TEDC2
THAP7
THEMIS2
TINAGL1
TLE5
TMEFF2
TMEM150A
TMEM231
TMEM41A
TMIE
TNIP3
TNK2
TNP2
TNS2
TRAPPC14
TRIM27
TRIM42
TROAP
TRPV6
TSPAN4
TTPA
TXNDC5
UBQLN4
UTP23
UXT
VASN
WDR25
WT1-AS
XCL2
YIPF3
ZDHHC1
ZFTRAF1
ZFYVE21
ZNF124
ZNF319
ZNF32
ZNF330
ZNF408
ZNF414
ZNF417
ZNF439
ZNF440
ZNF446
ZNF497
ZNF580
ZNF581
ZNF587
ZNF648
ZNF672
ZNF688
ZNF786
ZNF837
|
| Entrez ID |
1621 |
388677 |
| HPRD ID |
01963
|
14833
|
| Ensembl ID |
ENSG00000123454
|
ENSG00000264343
|
| Uniprot IDs |
P09172
|
A0A8I5KR58
Q7Z3S9
|
| PDB IDs |
4ZEL
|
Not available
|
| GO Terms Enriched among Interactors |
|
|