A reference catalog of DNA palindromes in the human genome and their variations in 1000 Genomes
Madhavi K. Ganapathiraju, Sandeep Subramanian, Srilakshmi Chaparala and Kalyani B. Karunakaran.Human Genome Variation 7, 40 (2020). https://doi.org/10.1038/s41439-020-00127-5
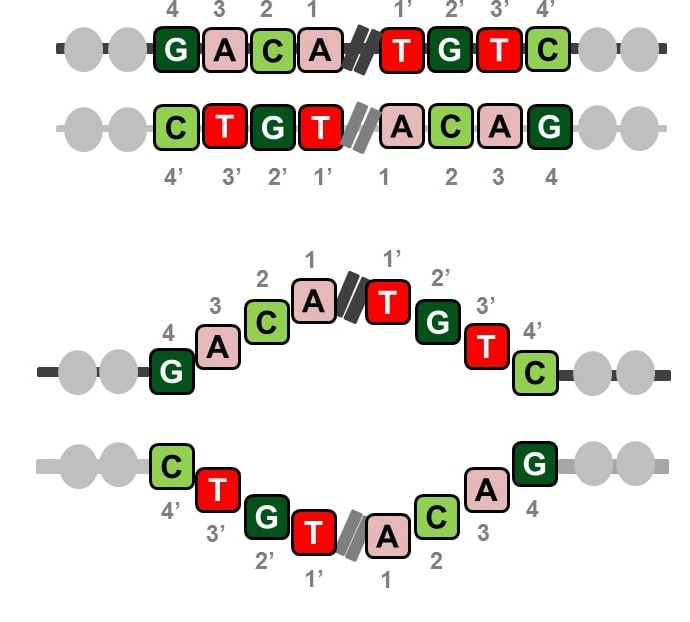
Abstract: A palindrome in DNA is like a palindrome in language, but when read backwards, it is a complement of the forward sequence; effectively, the two halves of a sequence complement each other from its midpoint like in a double strand of DNA. Palindromes are distributed throughout the human genome and play significant roles in gene expression and regulation. Palindromic mutations are linked to many human diseases, such as neuronal disorders, mental retardation, and various cancers. In this work, we computed and analyzed the palindromic sequences in the human genome and studied their conservation in personal genomes using 1000 Genomes data. We found that ~30% of the palindromes exhibit variation, some of which are caused by rare variants. The analysis of disease/trait-associated single-nucleotide polymorphisms in palindromic regions showed that disease-associated risk variants are 14 times more likely to be present in palindromic regions than in other regions. The catalog of palindromes in the reference genome and 1000 Genomes is being made available here with details on their variations in each individual genome to serve as a resource for future and retrospective whole-genome studies identifying statistically significant palindrome variations associated with diseases or traits and their roles in disease mechanisms.
Full Catalog (5 GB tar.gz file) (Supplementary File 3) of palindromes in reference genome and their variations in 1000 Genomes. The README file has all the necessary information, but can be improved! We will do that by end of December. Write to Madhavi if you need files for individual chromosomes or any other related information.